How genetic and molecular innovation in pathology will improve NHS outcomes for patients
The Genetic Revolution
The march of the genetic revolution has seen the understanding of the human genome grow at an exponential rate, surpassing traditional development time lines. Diagnostic and pharmaceutical companies are having to play catch-up with the opportunities and benefits this area of medicine has to offer patients, clinicians and the wider population.
The diagnostic paradigm is set to change, no longer will genetic testing be the final confirmatory test of a proposed diagnostic hypothesis, that has been shaped by countless other traditional pathology tests, but the starting point. It will become the basis of a personalised treatment plan for that individual or even whole families from birth to head off a genetically described future disorder.
The Diagnostic Pathway: Now and then
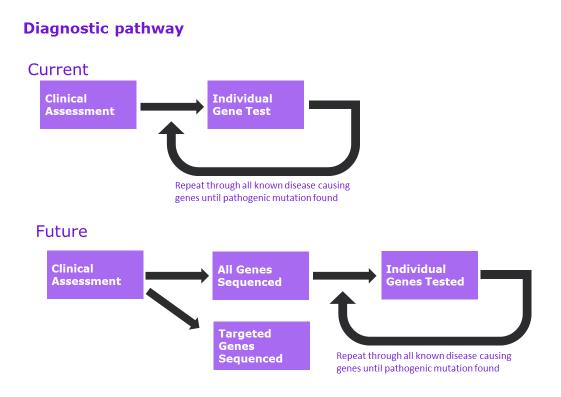
Current Advances
An example of the advances being made is the work being undertaken by Dr Tony Marinaki, Consultant Clinical Scientist in Viapath’s Purine Research Laboratory. Studying microRNAs (miRNA), which are small non-coding RNA molecules that are involved in regulation of gene expression, Dr Marinaki and his team showed in a pilot study that circulating levels of miR-132 were significantly decreased in patients with gestational diabetes mellitus at 24-30 weeks gestation compared to those women without gestational diabetes. These results raise the possibility of an alternative test to an oral glucose tolerance test for the diagnosis of gestational diabetes. The results also build upon the understanding of how gene expression can be linked to circulating markers for predicting disorders.
Personalised Medicine
As well as predicting and diagnosing a pathological disorder, treatment can be targeted. By building an understanding of the genetic make-up of an acquired genetic mutation, such as those seen in cancer, treatments can be personalised to ensure the best possible outcome for the patient, with the clinician being able to target specific monoclonal treatments against ligands, receptors and intracellular signalling components reducing the capacity of the cancer to spread. An example of this can be seen in the use of Herceptin in breast cancer in patients who express the HER2 genotype.
This scientific understanding expands the need for a more comprehensive diagnostic service, it also increases the need to ensure a complete diagnostic picture can be presented for each and every patient enabling best practice and targeted treatment plans that are appropriate, proportionate and with the optimum chance of success.
The anticancer drug 5-fluorouracil (5FU) is widely used for the treatment breast and colorectal cancer but is associated with severe toxicity in about 25% of patients. In work published in the British Journal of Cancer, researchers at Viapath showed that variation in dihydropyrimidine dehydrogenase (DPD), an enzyme involved in the degradation of pyrimidines, predicts severe toxicity to this therapy. On average, patients with a variant human DPD gene (DPYD) genotype spent seven days in hospital due to toxicity. Similarly, data presented at a recent National Cancer Research Conference by breast cancer oncologists from Guy’s Hospital showed that in a series of 44 patient, two patients experiencing toxicity were both DPD deficient. This illustrates that using pharmacogenetics to identify and manage at-risk patients will improve their quality of life, clinical outcomes and, even though diagnostic costs rise, the overall costs of patient pathways decrease.
As with all revolutionary steps in healthcare, there are other benefits that are being harnessed. It is not just the human genome that is of interest to healthcare scientists, but those of microbes. The rapid identification of an infective organism can directly improve the outcome for a patient. Basic point of care devices that can detect the DNA or RNA of a microbe are available. These enable the clinical team to target treatment faster, understand outbreaks and cohort patients in wards to reduce hospital acquired infections. Understanding the use and applicability of the technology is the challenge but one which will reap rewards. 
Innovation of this branch of pathology is not limited to a better understanding of the genetic basis of disease; it requires diagnostic and pathology providers, healthcare systems, industry and national programmes to build the infrastructure to support this. Much of the genetic revolution has not come from novel techniques, but a better method of handling significant quantities of data, to allow interpretation via clinically robust algorithm pipelines, to adopt and introduce the latest genetic sequencing technology and strategy. This can then be applied to deliver the patient focused approach that will be needed to realise fully the benefits. To put this into context, in recent years referrals for Comparative Genomic Hybridisation (CGH) array testing to Viapath’s Genetics laboratory at Guy’s Hospital showed that roughly 25% of the results revealed mutations that can lead to intellectual disability. Neurodevelopmental disorders cover a far bigger group than solely intellectual disability and Dr Ahn and his team have been trying to code the phenotype. These findings imply that targeting only one specific gene for neurodevelopmental disorders in the diagnosis of a patient is inefficient and more comprehensive testing strategies are needed to support a diagnosis. To date, Dr Ahn’s research has found 1421 genes with reported pathogenic variants associated with a neurodevelopmental phenotype. A collaboration with industry and funding from Viapath’s Innovation Academy’s Innovation Fund has allowed analysis using next generation sequencing of these genes to further understand the genetic mechanisms by which these conditions occur.
Advances in Technology
Providers are faced with a complex choice of technological approaches each with advantages and disadvantages depending upon the application. The use of HiSeq and Mieq high resolution techniques for high throughput genetic analysis for multiple genetic targets, whole exome and even whole genome, are able to accurately identify mutations in patients with congenital disorders. These are highly productive where the disorder can be characterised by thousands of different mutations to reach a positive diagnosis, but incredibly intensive in terms of the Healthcare Scientist’s time as they work through the sub-clinical and naturally occurring mutations. However, when looking for the presence or, indeed, absence of a single mutation or gene, this can be time consuming and costly for a single patient. More targeted technologies may prove to be a better option; such as Ion-Torrent that allows for a “load and go” operation, with all the required algorithms pre-loaded in the instrumentation. This approach is able to pick up a single gene rapidly, giving the clinician the additional information in time for inclusion into the histological report on a biopsy, increasing the support the pathology report has on the advanced management of the cancer patient.
The advances within the field of molecular diagnostics is also allowing the opportunity for new areas of diagnostics, particularly as the understanding of the genome increases the requirement to see the expressed and active molecules in identified mutations. This is giving rise to the clinical applications of epigenetics, proteomics and metabolomics amongst others.
These different technological approaches are costly and fundamentally different. Purchasing either at scale, ensuring a wide panel of instruments, close engagement with diagnostic instrument providers, guaranteeing the availability of appropriate genetic panels, in the case of Ion-Torrent, or purchasing access to dedicated high throughput sequencing lanes, together with an active development pipeline.
This allows the laboratory to remain responsive to clinical demands for both cancer and constitutional genetics. It is likely that diagnostic providers will need a range of these technologies to deliver the future advances in genetic medicine.
Another consideration that needs to be applied is information management: the transfer and storage of significantly sized files is going to impact upon many healthcare organisations. The size and scale that allows wide scale adoption and integration is on a scale with Radiology Picture Archiving and Communication System (PACS) systems and is likely to be linked into wider digital pathology solutions to enable efficiency of scale.
Adoption at scale across the UK is also limited by the available workforce. Bioinformatics as a separate healthcare science discipline is in its infancy, with the first cohort of nationally funded trainee scientists only part way through their registration training. Experience of existing scientists is concentrated into current large genetics centres, some equipped with strong organisational links that allow the broad adoption of molecular diagnostic techniques with others working in limited silos.
Large pathology providers, especially those that are able to direct their own strategy and investment choices, are well placed to take a national lead in the wide scale adoption of molecular techniques. The co-location of laboratories is not a pre-requisite, in fact allowing for centralisation builds upon the available limited expertise, enabling broader access to this essential knowledge base. Access to a cohort of experts, not just genetic experts, integrated into clinical teams is essential.
It is an often cited fact that technological adoption within the NHS takes on average 14 years. The demand and expectation for pathology departments to deliver a comprehensive molecular diagnostic service across all specialities is prevalent now, pressing even organisations investing heavily in next generation sequencing. In the current economic environment, adoption will come through partnerships with providers that operate at scale, with existing expertise and a proven clinical track record.
For further information, please contact
David Wells, Director of Operations, Reference Services
david [dot] wells [at] viapath [dot] co [dot] uk

