Genetic analysis of samples from miscarriages using a new efficient molecular testing strategy
Summary
Around 15% of pregnancies end in miscarriage; identifying the cause of pregnancy loss is important for couples and may be critical for the management of their future pregnancies.
The finding of chromosome imbalance in samples from miscarriages can provide a reason for pregnancy loss, may predict the risk of recurrence of miscarriage and inform reproductive options for couples.
Genetic investigation of recurrent miscarriage (defined as three or more miscarriages) is funded by the NHS.
Viapath’s combined QF-PCR/array CGH approach detects more abnormalities than previous techniques.
Why is it important to test samples from miscarriages?
Traditional testing of miscarriage products has involved culture of tissue, followed by G-banded chromosome analysis, a technique used to visualise the number and appearance of the cells’ chromosomes. However, this approach is labour-intensive, has a high failure rate (because tissue samples often do not grow in culture) and can only detect large imbalances of >10Mb in the chromosomes. Viapath’s Genetics Laboratory has replaced this methodology with a combined molecular approach using quantitative fluorescent polymerase chain reaction (QF-PCR) and array comparative genomic hybridisation (CGH) and have recently described the testing of more than 3,000 samples (Donaghue et al 2017).
Viapath’s new testing strategy
The new strategy involves two steps:
Step 1. QF-PCR
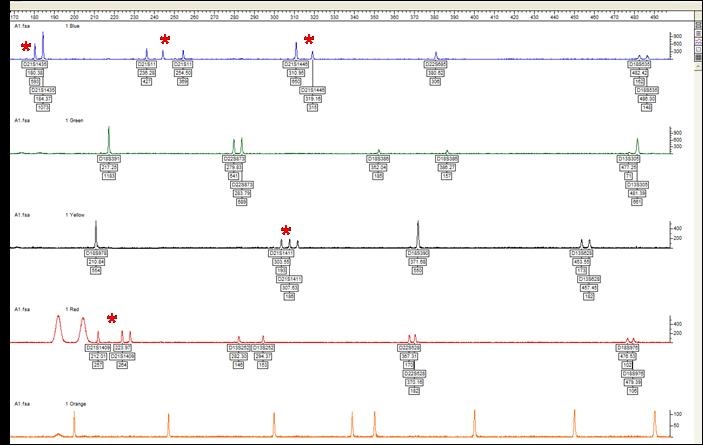
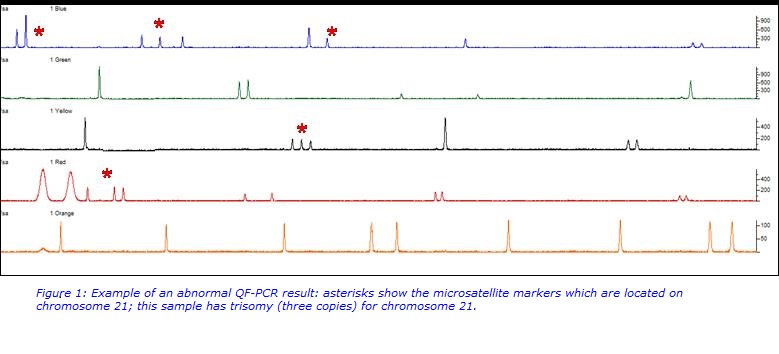
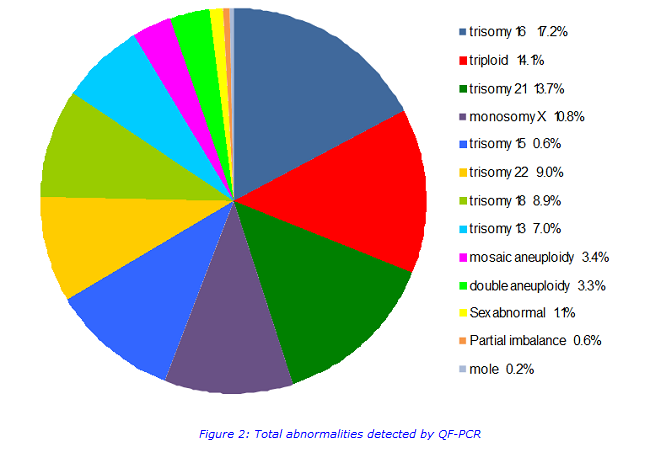
QF-PCR is a cost-effective, fast, semi-quantitative technique in which microsatellite regions of chromosomes are amplified by PCR. This assay will detect the most common chromosome abnormalities in miscarriage samples which are aneuploidy (an abnormal number of chromosomes in a cell) for chromosomes 13, 14, 15, 16, 18, 21, 22, X and Y. Examples of the abnormalities detected are trisomy (three copies) of chromosome 21 (Down syndrome), three copies of all chromosomes (triploid) or one copy of the X chromosome (Turner syndrome). All abnormalities detected by this assay are likely to have caused the miscarriage.
This assay detects an abnormality in approximately 25% of samples.
Step 2. array CGH
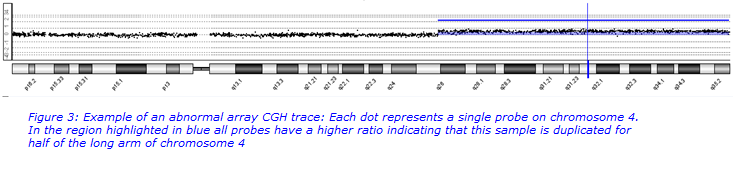
If no abnormality is detected on QF-PCR then array CGH testing is also carried out. This test will pick up more abnormalities than QF-PCR but it is more work-intensive. It scans for imbalances across the whole genome and detects imbalances >0.12Mb. All imbalances >1Mb are reported and any clinically significant imbalances which are <1Mb are reported. An abnormality is detected in approximately 10% of samples. Abnormalities include whole chromosome aneuploidies, unbalanced translocations, novel large deletions and duplications, small well known submicroscopic deletions and duplications and variants of unknown significance (VOUS).
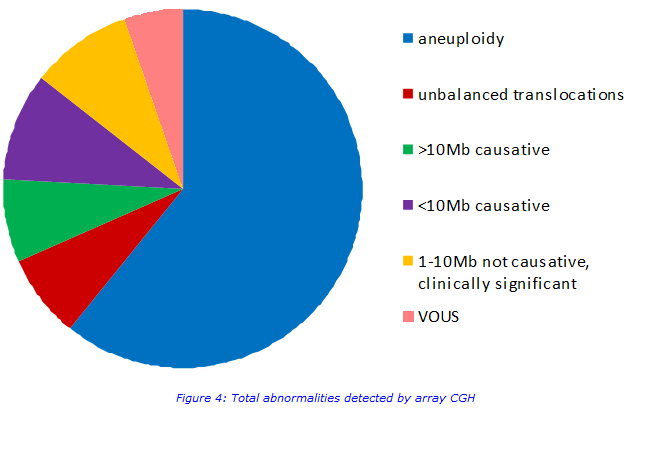
Imbalances which are not reported
All imbalances >1Mb are reported, however imbalances <1Mb are only reported if the imbalance has a known fully penetrant clinical phenotype. Variants of unknown significance (VOUS) are not reported; the clinical significance of these imbalances and any potential relevance to the miscarriage is currently unknown. Neurosusceptibility regions (which confer an increased risk of neurodisability) are also not reported; these imbalances will not have caused the miscarriage and their impact on the clinical features of any future children cannot be predicted.
Conclusion
This new strategy has a lower failure rate (1.4% compared to 30% for G-banded chromosome analysis) and reporting times are improved (89% of samples are reported within the target of 28 days compared to 79% for G-banded chromosome analysis). The new strategy also detects more imbalances, therefore the cause of the miscarriage is identified for more couples.
How can testing be obtained?
How can testing be obtained?
- Samples should be sent to Viapath’s Genetics Laboratory at Guys Hospital
- Samples are accepted from any centre. Testing is funded by the NHS if a patient has had three or more miscarriages; otherwise privately funded samples are accepted.
- Samples should be sent in a dry sterile container and ideally should arrive within 24 hours. Samples do not need to be shipped on ice but should NOT be formalin fixed.
Testing is usually completed within 28 days
Reference:
Donaghue C et al 2217. Efficient and cost-effective genetic analysis of products of conception and fetal tissues using a QF-PCR/array CGH strategy; five years of data. Mol Cytogenet.

